Hi, I'm Rakshit Kumar Jain.
A
Self-driven, quick starter, passionate programmer with a curious mind who enjoys solving a complex and challenging real-world problems.
About
I am a 4th Year Ph.D. Candidate in the Chemical and Biomolecular Engineering department at North Carolina State University. I enjoy problem-solving and coding geared towards molecular simulations. My doctorate work focuses on developing a new computational technique to study peptoids. I have worked on a number of physical systems including but not limited to peptides, nanogels, microgels and grain technologies. I have keen interest in using Molecular Thermodynamics and Statistical Mechanics to study a variety of real-world problems. After my doctoral studies, I hope to work as a postdoctoral scholar ultimately leading to a life as an academic because I find great pleasure in teaching and interacting with students.
- Languages: C, C++, Python, Fortran, Bash, HTML/CSS, Javascript
- Simulation Techniques: Discontinuous Molecular Dynamics, Molecular Dynamics, Ab initio Simulations, Monte Carlo, Kinetic Monte Carlo, Density Functional Theory, Discrete Element Method
- Libraries: GSL, BOOST, TRNG, NumPy, SciPy, Pandas, OpenCV
Looking for an opportunity to work in a challenging position combining my skills in simulations and Molecular Engineering, which provides professional development, interesting experiences and personal growth.
Publications
Luis Perez-Mas, Alberto Martin-Molina, Rakshit Kumar Jain, Manuel Quesada-Perez, Journal of Molecular Liquids, Vol. 288, 2019, 111101
Experience
- Tools: C, Visual Molecular Dynamics (VMD)
EFFECT OF DISPERSIVE FORCES BETWEEN NANOGELS AND THE IONS OF THE HOFMEISTER SERIES
Supervisor: Dr. Alberto Martin-Molina
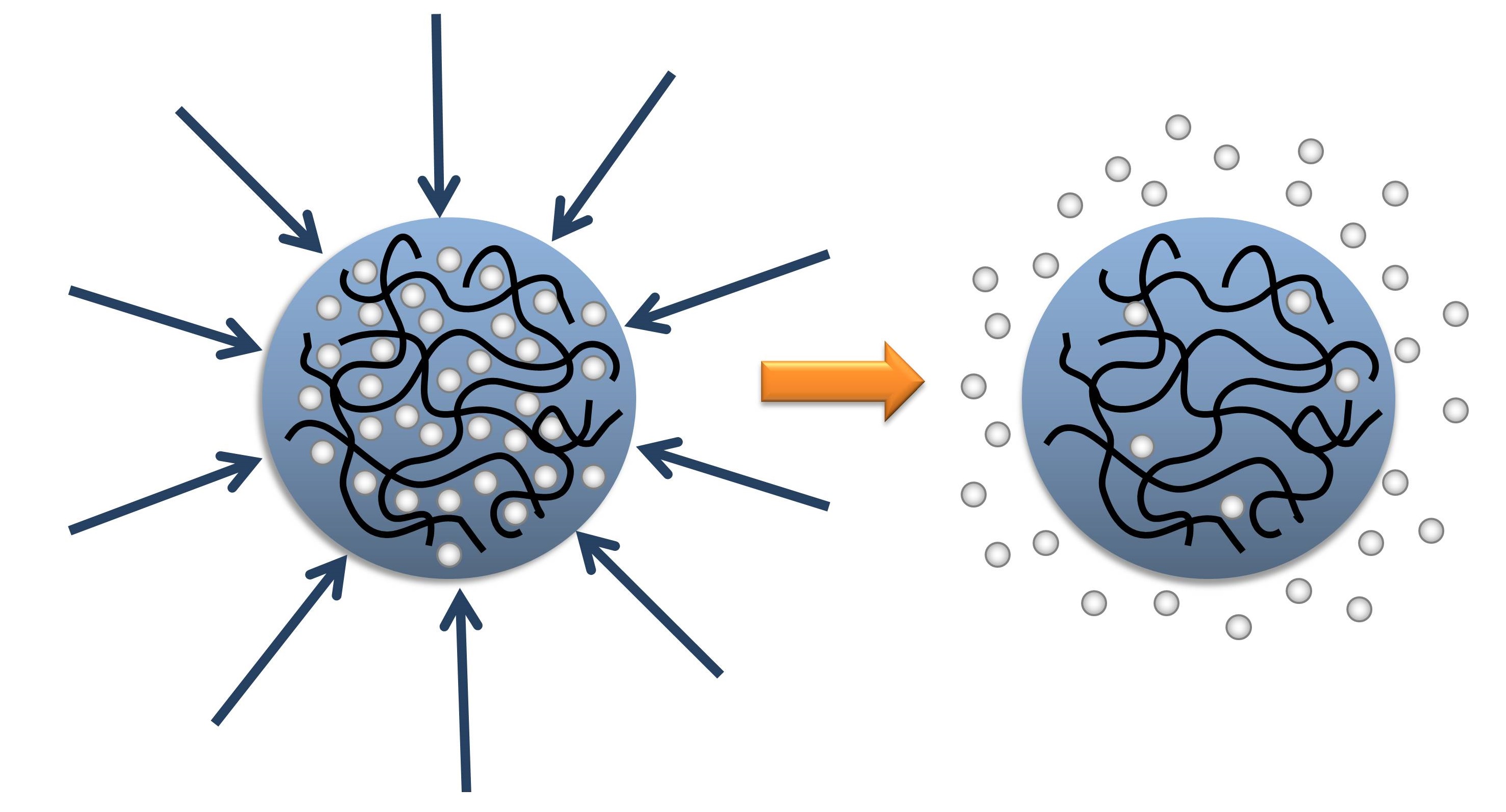
We studied poly-NIPAM, a polymer mesh important due to its specific thermalresponses. Electrostatic, hydrophilic and hydrophobic were the majorforces with the excluded volume interactions included using Lennard-Jones potential and dispersion forces. Parameters like ion distribution, the charge enclosed within the nanogel etc. were calculated with the view to perform a comparative study in the presence and absence of dispersion forces.
Research
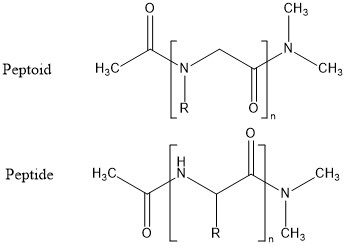
Dr. Erik E. Santiso, Dr. Carol Hall (North Carolina State University)
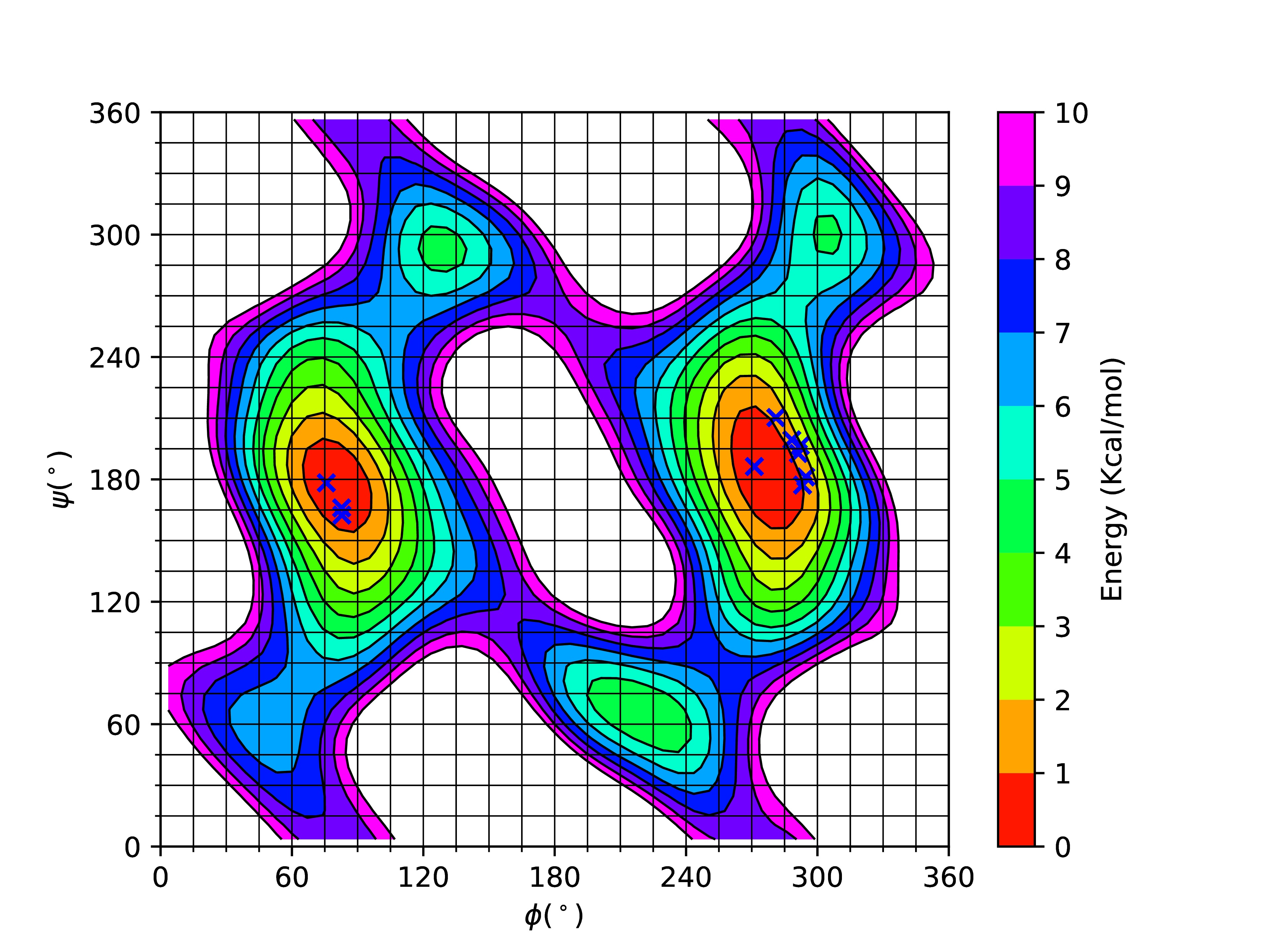
Dr. Erik E. Santiso, Dr. Carol Hall (North Carolina State University)
Dr. Prateek Jha (Indian Institute of Technology Roorkee)
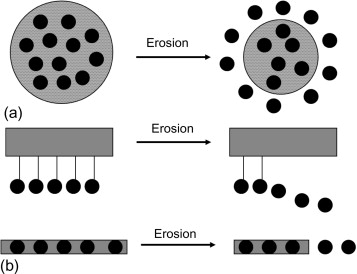
Dr. Prateek Jha (Indian Institute of Technology Roorkee)
Here, modeled the behavior of carrier polymers and controlled release mechanisms under surface erosion due to a moving fluid, developing an improved model over the pre-existing ones and verifying the same with the help of dissolution experiments.

Dr. Anshu Anand (Indian Institute of Technology Roorkee)
Discrete Element Method (DEM) was used to model a fluidized bed to observe features under the effect of a gaseous medium from below. OpenFOAM was used to integrate fluid dynamics as LIGGGHTS can only simulate particle behavior.
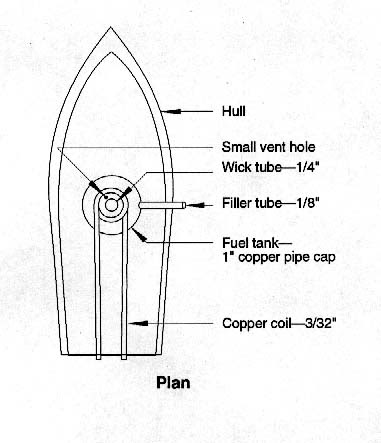
Dr. Gaurav Sharma (Indian Institute of Technology Roorkee)
A pop-pop boat moves when the boiler is filled with water, and then heated. There is no satisfactory model for the working of the boat and we aimed at finding a model to fit the experimental data using Bernoulli’s equation.
Teaching

Fall 2019-2021 (Dr. Peter Fedkiw, North Carolina State University)
Assisted as a TA for the course.

Spring 2019 (Dr. Erik Santiso, North Carolina State University)
Assisted as a TA for the course.
Skills
Languages
 C/C++
C/C++
 Python
Python
 Shell Scripting
Shell Scripting
 FORTRAN
FORTRAN
 HTML5
HTML5
 CSS3
CSS3
Softwares
 NAMD
NAMD
 Gaussian13
Gaussian13
 MATLAB
MATLAB
 LIGGGHTS
LIGGGHTS
 ANSYS FLUENT
ANSYS FLUENT
 LATEX
LATEX
Simulation Techniques
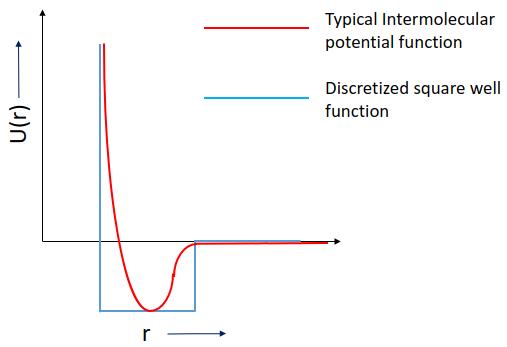 Discontinuous Molecular Dynamics
Discontinuous Molecular Dynamics
 Molecular Dynamics
Molecular Dynamics
 Ab Initio Methods
Ab Initio Methods
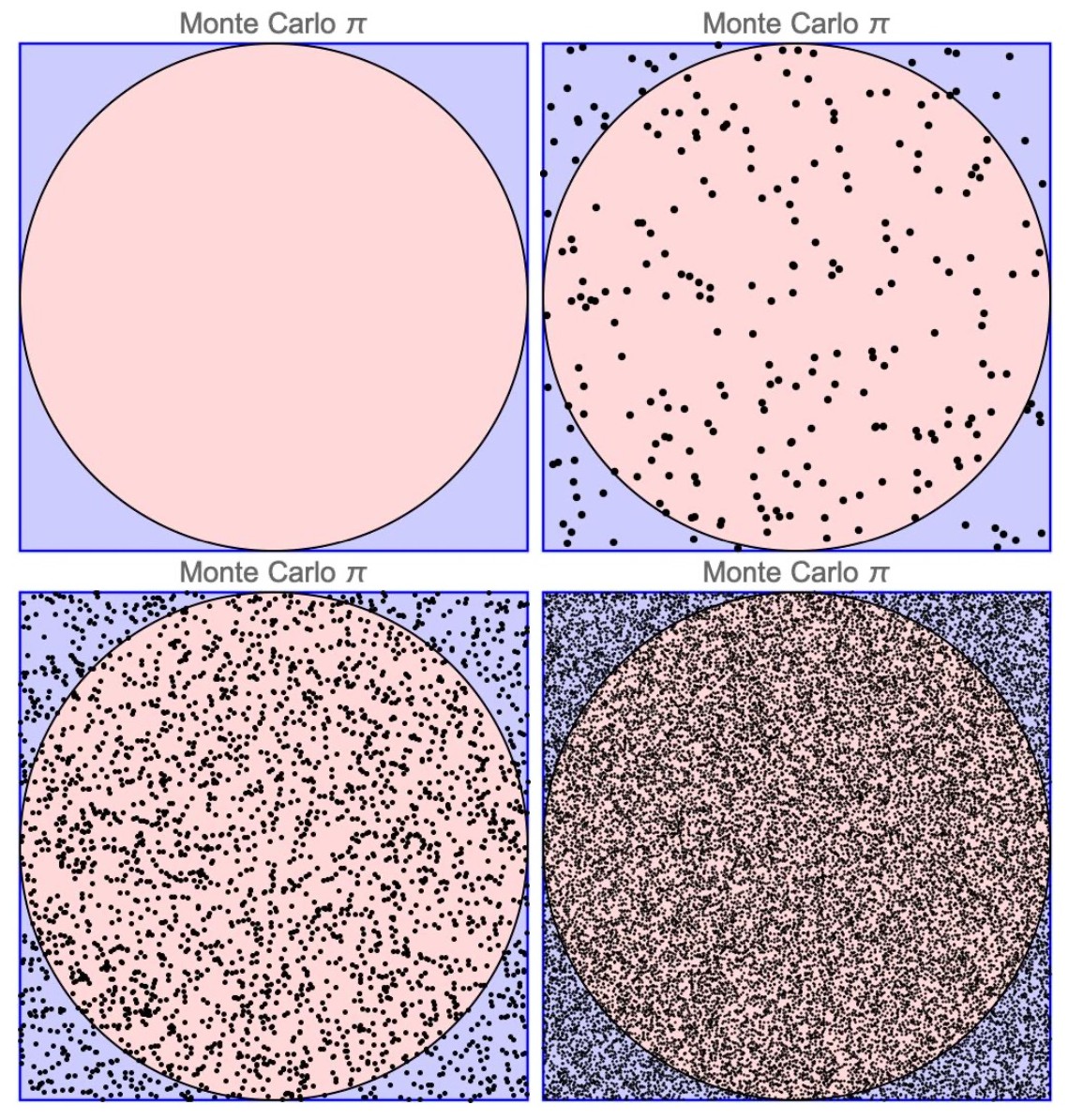 Monte Carlo
Monte Carlo
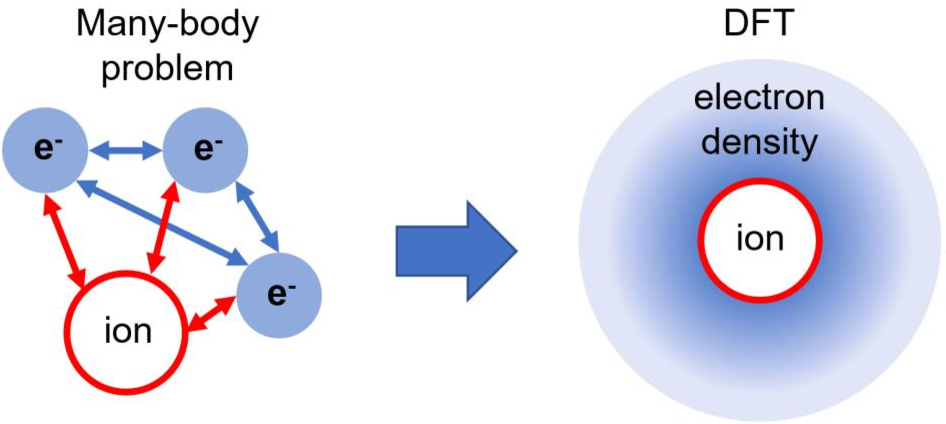 Density Functional Theory
Density Functional Theory
 Discrete Element Methods
Discrete Element Methods
Education
North Carolina State University
Raleigh, NC, USA
Degree: Ph.D. in Chemical and Biomolecular Engineering
CGPA: 4.0/4.0
- Special Topics "Introduction to Molecular Simulations"
- Statistical Physics
- Multiscale Modeling of Matter
- Chemical Engineering Process Modeling
Relevant Courseworks:
Indian Institute of Technology Roorkee
Roorkee, India
Degree: Bachelor of Technology in Chemical Engineering
CGPA: 3.56/4
- Engineering Analysis and Process Modeling
- Computer Programming and Numerical Methods
- Computational Fluid Dynamics
- Transport Phenomena
- Thermodynamics and Chemical Kinetics
Relevant Courseworks: